Discover the new GENOVESA website!
Visit genovesa.biovendor.group to learn more about GENOVESA software, explore features, and request a demo. Simplify your data management with GENOVESA today!
Type
Bioinformatic analysis software
Description
The analysis of genetic NGS data can be a daunting task that often requires specialized computing skills by the user. We have developed our proprietary web-based bioinformatic analysis software – GENOVESA as an effortless solution for everyone.
GENOVESA is intuitive and easy to use. For beginners in NGS analysis, it allows a quick and easy generation of relevant information of their NGS data, without the need of their own dedicated bioinformatic experts. On the other hand, GENOVESA allows experienced users to adjust and modify parameters, use complex filtering rules, and visually evaluate variants at the individual reading level.
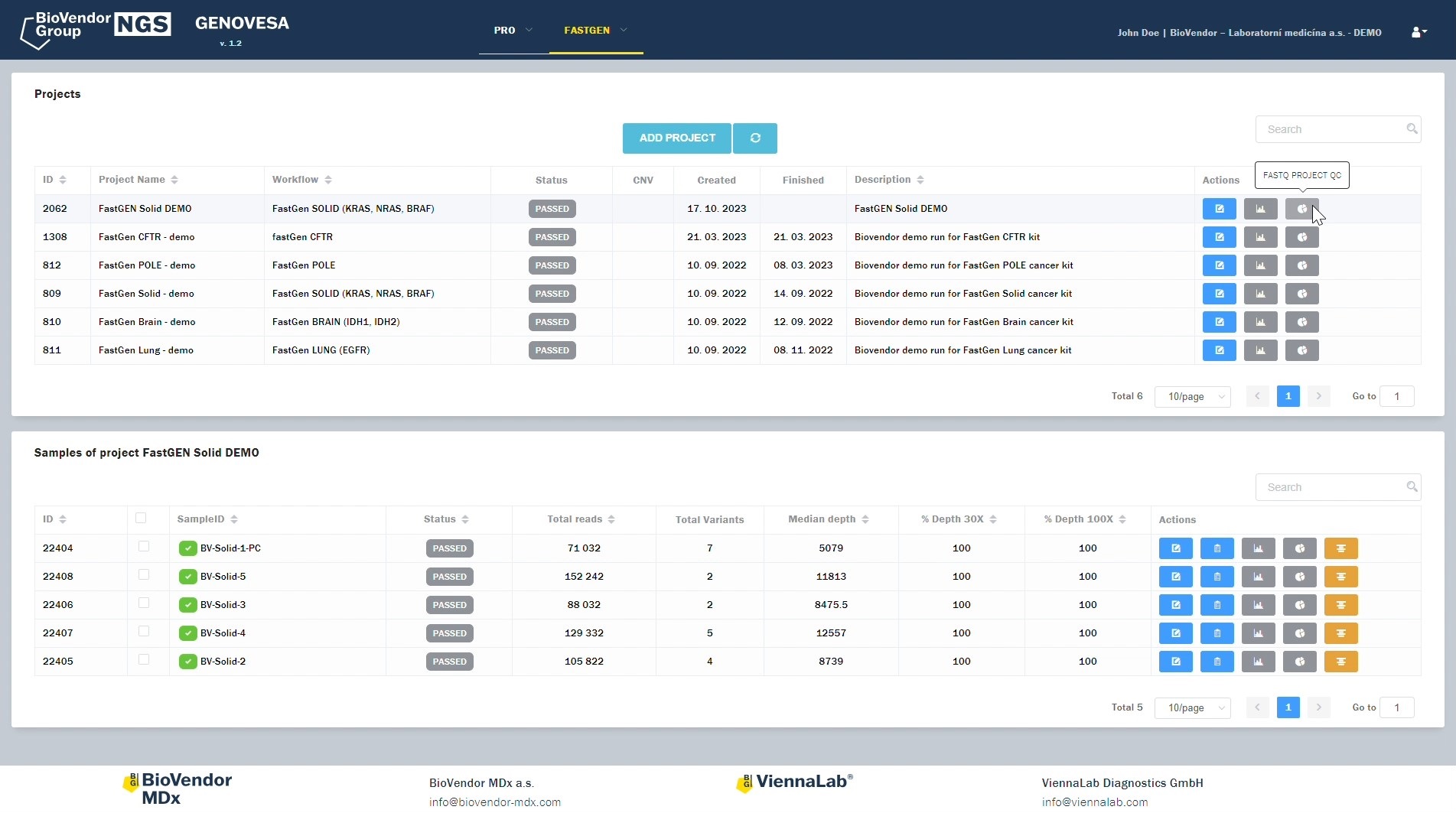
GENOVESA is an essential part of the ViennaLab NGS Assays and is included with every kit purchase, such as the Hereditary Cancer NGS Assay, the Somatic Mutations NGS Assay and the CES NGS Assay.
GENOVESA is an essential part of the fastGEN kits and is included with every kit purchase, such as fastGEN Solid Cancer kit, fastGEN Lung Cancer kit, fastGEN Brain Cancer kit and fastGEN POLE Cancer kit and so on.
GENOVESA is a cloud-based Next-Generation Sequencing (NGS) software that prioritizes data security through HTTPS encryption. We do not rely on third-party providers like Google or Amazon; instead, we host your data in our secure data center located in Prague, Czech Republic. Rest assured, we strictly protect patient sequencing data. We also do not collect any sensitive patient data - in GENOVESA it is possible upload only FASTQ files.
Key features
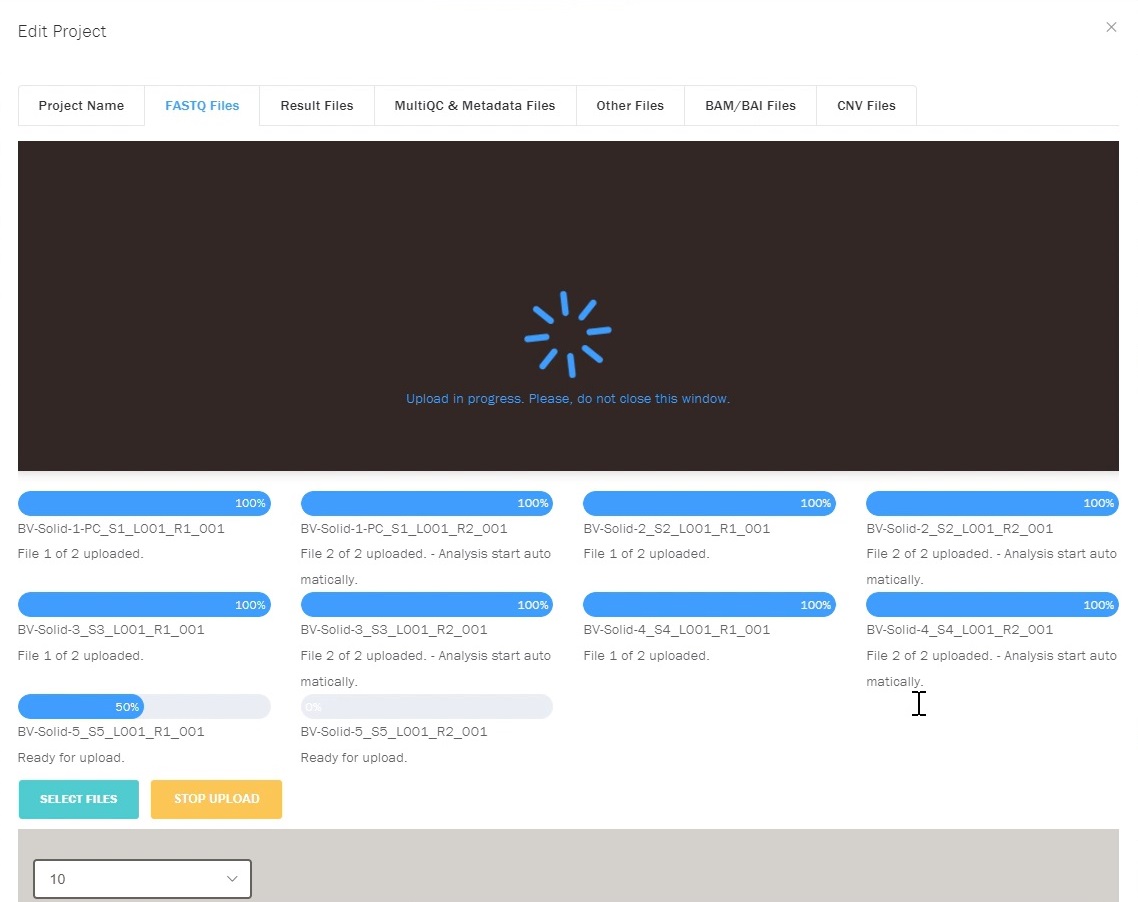
- Uploading of fastq files through a secure web interface.
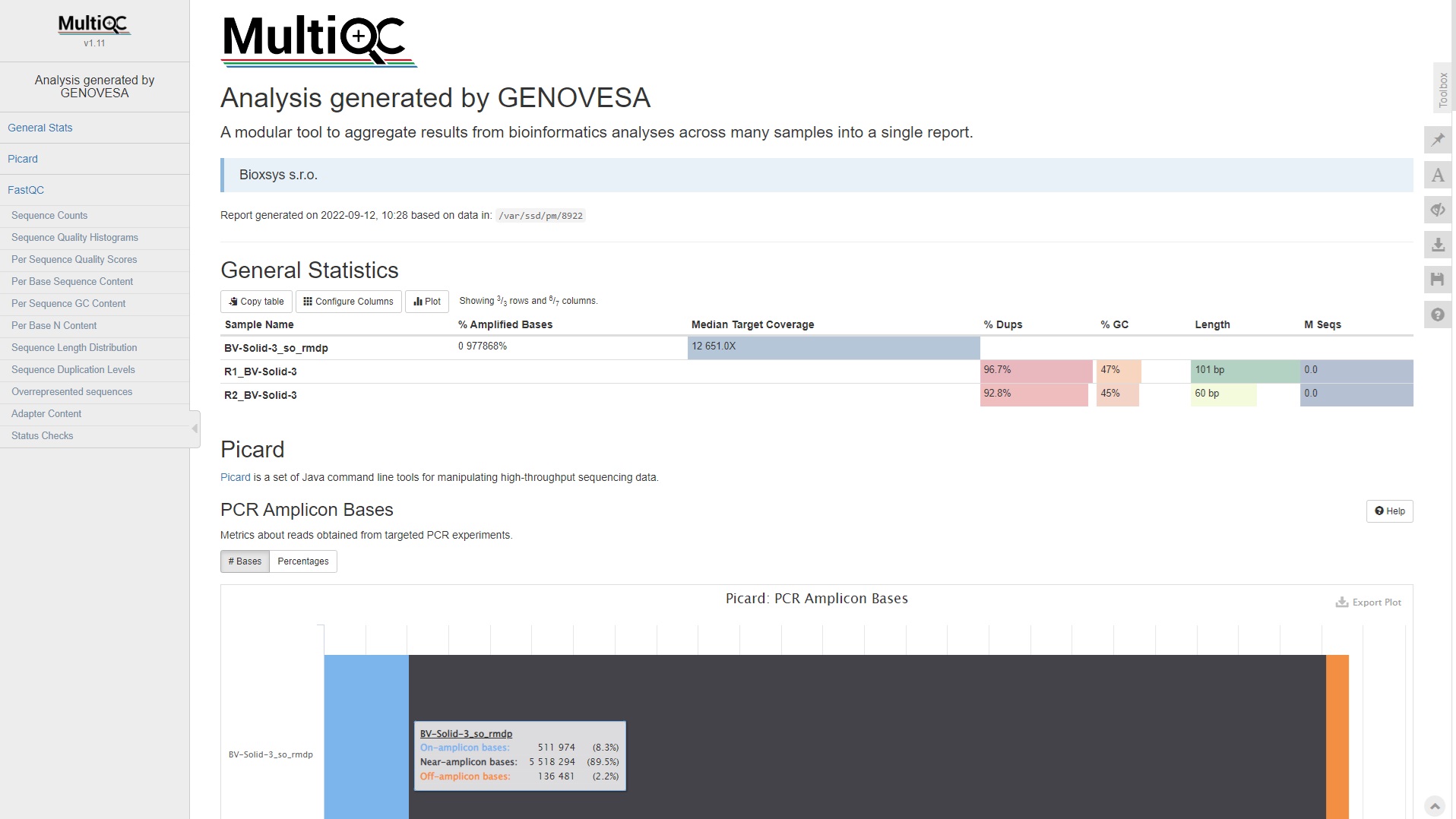
- Automated processing and quality control metrics of sequencing data.
- Identification and annotation of variants from up-to-date relevant databases, as well as selection of variants for the generation of a meaningful report.
- ViennaLab preset filters for simplified and time-saving identification of clinically relevant variants without the need to look for the needle in the haystack.
- Custom filter section for tailoring parameters to specific customer needs (e.g. maximum population frequency, minimum read depth, ClinVar pathogenicity classification).
- Visual display and inspection of SNVs and InDels is possible through a seamlessly integrated viewer. CNV and structural variants (SV) analysis and visualization are available upon request.
Note
GENOVESA - The cloud-based database bioinformatic and interpretation system