Type
Recombinant protein
Description
Recombinant SARS-CoV-2 Spike protein N-terminal domain (NTD). This domain shows the lowest sequence ID compared to SARS-CoV Spike protein. The protein is produced in HEK293 cells and purified from culture supernatant by Protein G chromatography.Contains Spike protein 1-260, C-terminally tagged with sheep Fc.
Source
HEK293
Purity
95%
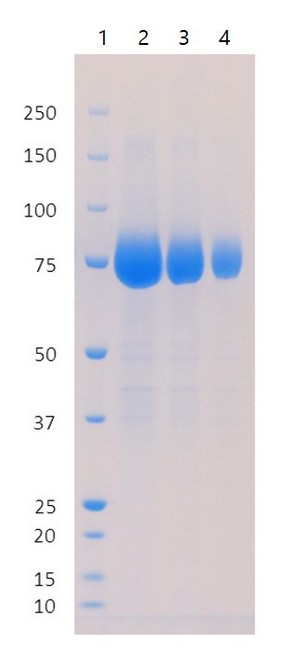
SDS-PAGE Gel
SDS-PAGE: REC31835 run on a denaturing SDS-PAGE under reducing conditions, stained with Coomassie.
- MW marker
- Purified N-terminal domain (9 µg)
- Purified N-terminal domain (4.5 µg)
- Purified N-terminal domain (1.8 µg)
Formulation
Presented in Dulbecco’s phosphate buffered saline (DPBS).
Reconstitution
Defrost at ambient temperature
Applications
Western blotting, ELISA
Shipping
On ice. Upon receipt, store the product at the temperature recommended below.
Storage/Expiration
Store protein at -80°C. Protein remains stable until the expiry date when stored at -80°C. Avoid repeated freezing/thawing cycles.
Note
This product is intended for research use only.
Research topic
Immune Response, Infection and Inflammation, COVID-19
Summary
Coronaviruses (CoVs), within the order Nidovirales, are enveloped, single-strand, positive-sense RNA viruses with a large genome of approximately 30 kbp in length. A human infecting coronavirus (viral pneumonia) initially known as 2019 novel coronavirus (2019-nCoV) was found in the fish market at the city of Wuhan, Hubei province of China in December 2019. The virus is now named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
SARS-CoV-2 shares an 87% identity to the 2 bat-derived severe acute respiratory syndrome 2018 SARS-CoV-2 located in Zhoushan of eastern China. SARS-CoV-2 has an analogous receptor-BD-structure to that of 2018 SARS-CoV, even though there is a.a. diversity so thus the SARS-CoV-2 might bind to ACE2 receptor protein (angiotensin-converting enzyme 2) in humans.
While bats are possibly the host of SARS-CoV-2, researchers suspect that animal from the ocean sold at the seafood market was an intermediate host. RSCU analysis proposes that the SARS-CoV-2 is a recombinant within the viral spike glycoprotein between the bat coronavirus and an unknown coronavirus.
Coronaviruses contain at least four structural proteins: Spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein.
The spike (S) glycoprotein is a type I transmembrane glycoprotein that plays an important role in mediating viral infection and is common to all HCoVs. The S proteins consist of two subunits, S1 and S2. The S1 subunit binds the cellular receptor through its receptor-binding domain (RBD), followed by conformational changes in the S2 subunit, which allows the fusion peptide to insert into the host target cell membrane. The heptad repeat 1 (HR1) region in the S2 subunit forms a homotrimeric assembly, which exposes three highly conserved hydrophobic grooves on the surface that bind heptad repeat 2 (HR2). This six-helix bundle (6-HB) core structure is formed during the fusion process and helps bring the viral and cellular membranes into close proximity for viral fusion and entry. Thus, the S protein is an important target protein for the development of specific drugs.