COVID-19 is a world-wide emergency with the first cases reported in December 2019 and more than 696,000 deaths and 18,000,000 cases of SARS-CoV-2 infection have been reported globally as of August 5th (WHO).
Understanding adaptive immunity to SARS-CoV2 is important for vaccine development, interpreting coronavirus disease 2019 (COVID-19) pathogenesis, and calibration of pandemic control measures.
An understanding of human T cell responses to SARS-CoV-2 is lacking, due to the rapid emergence of the pandemic. There is an urgent need for foundational information about T cell responses to this virus.
The first steps for such an understanding are the ability to quantify the virus-specific CD4+ and CD8+ T cells. Such knowledge is of immediate relevance, as it will provide insights into immunity and pathogenesis of SARS-CoV-2 infection, and the same knowledge will assist vaccine design and evaluation of candidate vaccines. Estimations of immunity are also central to epidemiological model calibration of future social distancing pandemic control measures.
Such projections are dramatically different depending on whether SARS-CoV-2 infection creates substantial immunity, and whether any crossreactive immunity exists between SARS-CoV-2 and circulating seasonal ‘common cold’ human coronaviruses. Definition and assessment of human antigen-specific SARS-CoV-2 T cell responses are best made with direct ex vivo T cell assays using broad-based epitope pools and assays capable of detecting T cells of any cytokine polarization.
There is also great uncertainty about whether adaptive immune responses to SARS-CoV2 are protective or pathogenic, or whether both scenarios can occur depending on timing, composition, or magnitude of the adaptive immune response. Hypotheses range the full gamut based on available clinical data from severe acute respiratory disease syndrome (SARS) or MERS or animal model data with SARS in mice, SARS in NHPs or FIPV in cats.
Based on data from SARS patients in 2003-2004 (caused by SARS-CoV, the most closely related human betacoronavirus to SARS-CoV-2), and based on the fact that most acute viral infections result in development of protective immunity, a likely possibility has been that substantial CD4+ T cell, CD8+ T cell, and neutralizing antibody responses develop to SARS-CoV-2 and all contribute to clearance of the acute infection; and, as a corollary, some of the T and B cells are retained long-term (i.e., multiple years) as immunological memory and protective immunity against SARS-CoV2 infection.
However, a contrarian viewpoint is also legitimate. While most acute infections result in the development of protective immunity, available data for human coronaviruses suggest the possibility that substantive adaptive immune responses can fail to occur and robust protective immunity can fail to develop. A failure to develop protective immunity could occur due to a T cell and/or antibody response of insufficient magnitude or durability, with the neutralizing antibody response being dependent on the CD4+ T cell response.
Thus, there is urgent need to understand the magnitude and composition of the human CD4+ and CD8+ T cell responses to SARS-CoV-2. If natural infection with SARS-CoV-2 elicits potent CD4+ and CD8+ T cell responses commonly associated with protective antiviral immunity, COVID-19 is a strong candidate for rapid vaccine development.
Immunopathogenesis in COVID-19 is a serious concern. It is most likely that an early CD4+ and CD8+ T cell response against SARS-CoV-2 is protective, but an early response is difficult to generate because of efficient innate immune evasion mechanisms of SARS-CoV-2 in humans. Immune evasion by SARS-CoV-2 is likely exacerbated by reduced myeloid cell antigen presenting cell (APC) function or availability in the elderly. In such cases, it is conceivable that late T cell responses may instead amplify pathogenic inflammatory outcomes in the presence of sustained high viral loadsin the lungs, by multiple hypothetical possible mechanisms. Critical (ICU) and fatal COVID-19 (and SARS) outcomes are associated with elevated levels of inflammatory cytokines and chemokines, including IL-6.
Vaccine developmentagainst acute viral infections classically focuses on vaccine-elicited recapitulation of the type of protective immune response elicited by natural infection.
Knowledge of SARS-CoV-2 proteins and epitopes recognized by human T cell responses is of immediate relevance, as it will allow for monitoring of COVID-19 immune responses in laboratories worldwide. Epitope knowledge will also assist candidate vaccine design and facilitate evaluation of vaccine candidate immunogenicity. Almost all of the current COVID-19 vaccine candidates are focused on the spike protein.
The results of these recent study demonstrated that the epitope MPs are reagents well suited to analyze and detect SARS-CoV-2-specific T cell responses with limited sample material. The researchers also developed and tested peptide pools corresponding to each of the 25 proteins encoded in the SARS-CoV2 genome.
An optimal vaccine CD8+ T cell response to SARS-CoV-2 might benefit from additional class I epitopes, such as the ones derived from the M,nsp6, ORF3a and or N.
More studies are required, but the data here appear to predominantly represent a classical TH1 response to SARS-CoV-2.
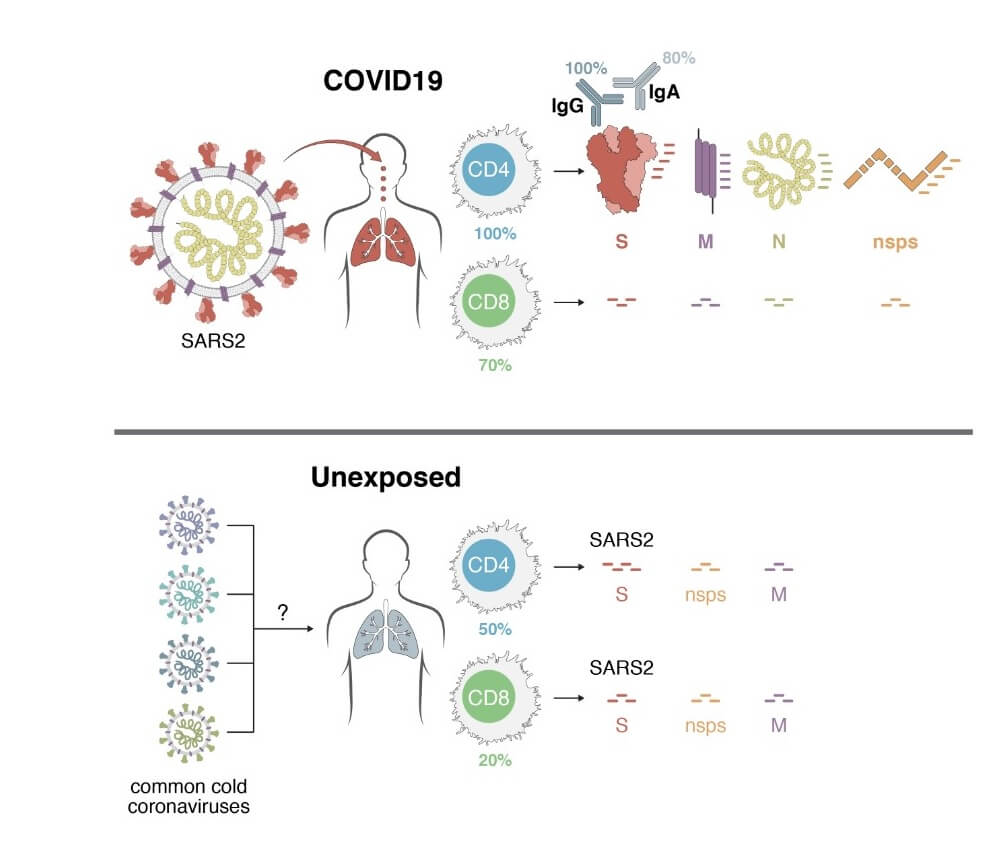
Using HLA class I and II predicted peptide ‘megapools’, circulating SARS-CoV-2−specific CD8+ and CD4+ T cells were identified in ~70% and 100% of COVID-19 convalescent patients, respectively.
CD4+ T cell responses to spike, the main target of most vaccine efforts, were robust and correlated with the magnitude of the anti-SARS-CoV-2 IgG and IgA titers.
Importantly, SARS-CoV-2−reactive CD4+ T cells were detected in ~40-60% of unexposed individuals, suggesting cross-reactive T cell recognition between circulating ‘common cold’ coronaviruses and SARS-CoV-2.
